[ad_1]
Radiology is a crucial part of diagnosing and treating illness by medical imaging procedures corresponding to X-rays, computed tomography (CT), magnetic resonance imaging (MRI), nuclear medication, positron emission tomography (PET) and ultrasound. The everyday radiology workflow entails handbook steps, notably across the protocoling course of. With Massive Language Fashions (LLMs), we will automate a few of this administrative burden.
Present State: Radiology Workflows
To discover in additional element, let’s dive into the everyday radiology workflow. Initially, a affected person might go to their supplier, reporting lingering results from a latest concussion. The supplier compiles the affected person’s notes into their Digital Well being File (EHR) and requests imaging, corresponding to a CT scan. Subsequently, a radiologist evaluations the medical notes and assigns an acceptable protocol label to the order, corresponding to ‘CT of Mind with distinction.’ This label guides the imaging technician in executing the order, resulting in the examination and subsequent evaluate of outcomes.

Figuring out Administrative Burden in Radiology Workflows
Now, why is the handbook strategy of assigning protocol labels important? There are two major causes.
Firstly, to mitigate inaccuracies inherent in human error. Our goal is not to interchange radiologists, however to reinforce their decision-making course of, growing consistency and lowering errors.
Secondly, radiologists, being among the many highest-paid medical professionals, spend roughly 3.5 – 6.2%(1,2) of their time, roughly equal to $17,000 to 30,000 yearly, on label assignments. By optimizing this course of, radiologists can redirect their efforts towards extra impactful duties.
Addressing Guide Duties with Massive Language Fashions (LLMs)
How did we tackle this problem? Initially, we generated an artificial dataset utilizing ChatGPT, mimicking supplier notes and corresponding protocol labels. Whereas not advisable for buyer use resulting from authorized constraints, it successfully helps this proof of idea.
We then chosen Meditron-7b because the foundational LLM for fine-tuning. Meditron, an open-source LLM skilled on PubMed articles, is apt for healthcare and life sciences functions, and nonetheless might be improved with fine-tuning. Basis fashions like Meditron are broad, and our use case calls for specificity. Nice-tuning tailors the mannequin to our specialised necessities round radiology protocoling.
To make sure cost-effectiveness, we applied Parameter Environment friendly Nice Tuning (PEFT). This strategy entails freezing a subset of parameters throughout the fine-tuning course of, thereby optimizing solely a fraction of parameters. By way of decomposition of the parameter matrix, we markedly cut back computational necessities whereas preserving efficiency. For example, think about a matrix (W) representing pretrained weights with dimensions of 10,000 rows by 20,000 columns, yielding a complete of 200 million parameters for updating in typical fine-tuning. With PEFT, we decompose this matrix into two smaller matrices utilizing an internal dimension hyperparameter. For instance, by decomposing the W matrix into matrices A (10,000 rows by 8 columns) and B (8 rows by 20,000 columns), we solely must replace 240,000 parameters. This signifies an approximate 99% discount within the variety of parameters requiring updates!
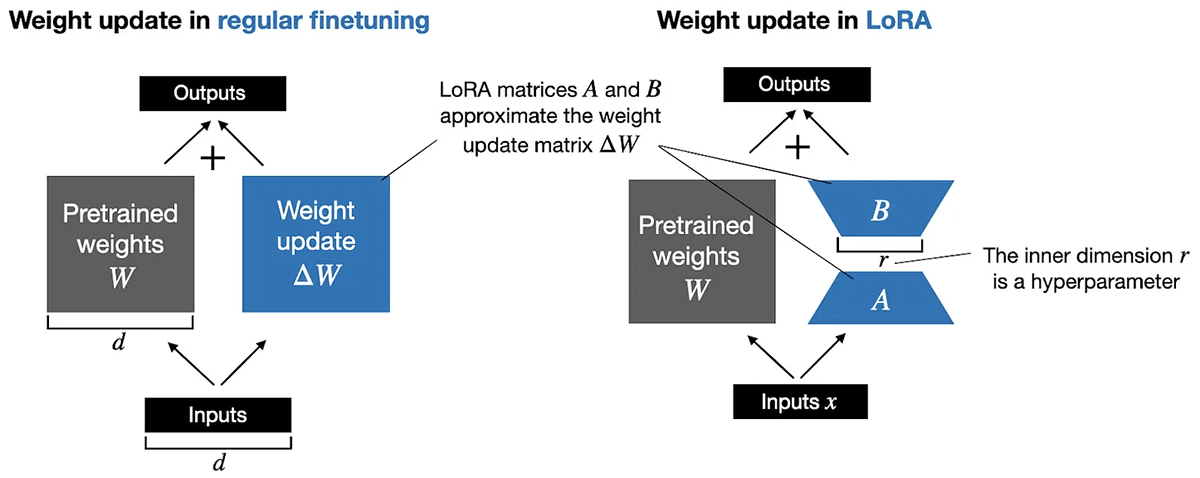
We applied QLoRA (Quantized Decrease Rank Adaption) as a part of our PEFT technique. QLoRA is a 4-bit transformer that integrates high-precision computing with a low-precision storage strategy. This ensures the mannequin stays compact whereas sustaining excessive efficiency and accuracy ranges.
QLoRA introduces three modern ideas aimed toward lowering reminiscence utilization whereas sustaining high-quality efficiency: 4-bit Regular Float, Double Quantization, and Paged Optimizers.
- 4-bit Regular Float: This novel information sort facilitates freezing subsets of neural networks, enabling Parameter Environment friendly Nice Tuning (PEFT) to attain efficiency ranges similar to 16-bit precision. By back-propagating gradients by a frozen 4-bit pre-trained Language and Studying Mannequin (LLM) to LoRA, the burden matrix is decomposed based mostly on the provided rank parameter (r) throughout the fine-tuning course of. Decrease values of r yield smaller replace matrices with fewer trainable parameters.
- Double Quantization: This course of entails mapping mannequin weights to decrease bit depths to scale back reminiscence and computational necessities. For example, beginning with a 16-bit NF, it may be quantized to eight bits, adopted by a subsequent quantization to 4 bits. This technique, utilized twice, leads to important reminiscence financial savings, roughly 0.37 bits per parameter on common, equal to roughly 3GB for a 65B mannequin (4).
- Paged Optimizers: Leveraging the idea of a “pager,” which manages information switch between major reminiscence and secondary storage, Paged Optimizers optimize the association of mannequin weights and biases in reminiscence. This optimization minimizes the time and sources required, notably throughout the processing of mini-batches with prolonged sequence lengths. By stopping reminiscence spikes related to gradient checkpointing, Paged Optimizers mitigate out-of-memory errors, traditionally hindering fine-tuning giant fashions on a single machine.
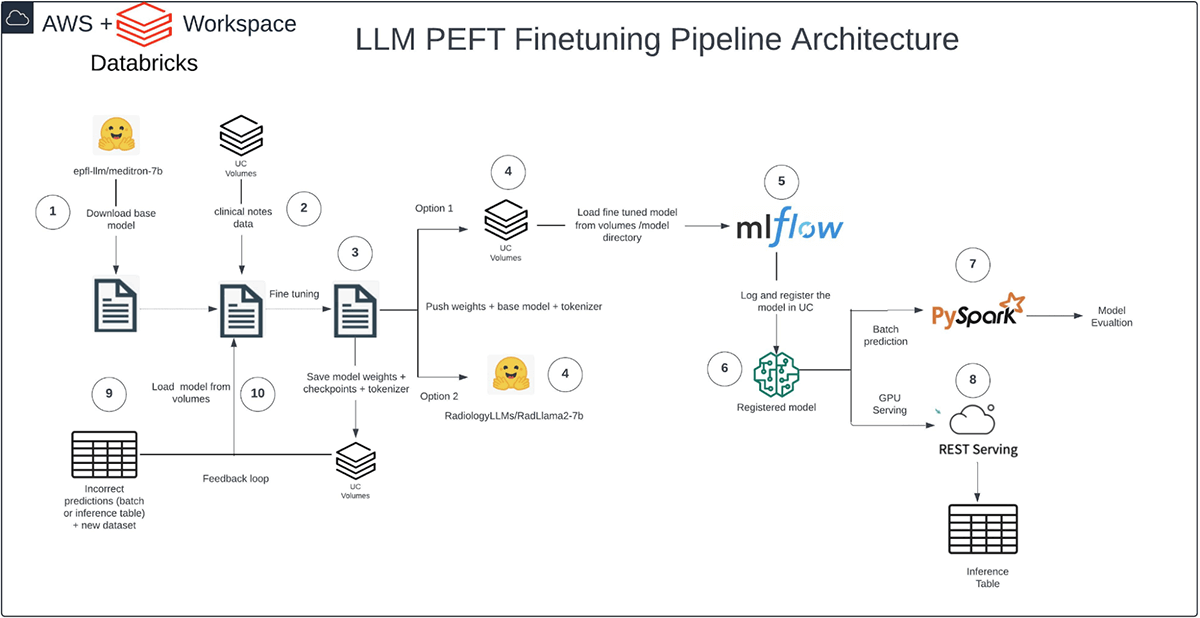
Our pipeline runs solely on Databricks and follows a structured movement: We acquired the bottom Meditron mannequin from Hugging Face into Databricks, imported our artificial dataset into Unity Catalog (UC) Volumes, and executed the fine-tuning course of with PEFT QLoRA in Databricks.
We saved mannequin weights, checkpoints, and tokenizer in UC Volumes, logged the mannequin with MLflow, and registered it in UC. Lastly, we utilized PySpark for inference with batch prediction. Semantic similarity served as our analysis metric, contemplating context within the analysis course of and making certain versatility throughout textual content era and summarization duties.
The advantage of executing this pipeline inside Databricks lies in its complete workflow capabilities and built-in governance throughout all layers. For example, we will prohibit entry to delicate affected person notes authored by physicians, making certain that solely approved personnel can view them. Equally, we will management the utilization of mannequin outputs, making certain that they’re accessed solely by related healthcare professionals, corresponding to radiologists.
Furthermore, it is noteworthy that every one options utilized on this setup are typically accessible, making certain compliance with HIPAA rules.
We belief that our prospects will uncover this pipeline to be a priceless asset for optimizing reminiscence utilization throughout varied fine-tuning situations in healthcare and life sciences.
Image this: the subsequent time a radiologist evaluations a doctor’s notes, detailing complicated medical historical past and diagnoses—corresponding to ‘Historical past: coronary heart failure, hepatic vein; Analysis: concern for liver laceration post-procedure, post-biopsy, on apixaban’—our system ensures the correct task of the suitable protocol label, like ‘CT Stomach W Distinction,’ moderately than ‘CT Mind.’ This precision can considerably improve diagnostic effectivity and affected person care.
Get began in your LLM use case with this Databricks Answer Accelerator.
References
(1) Dhanoa D, Dhesi TS, Burton KR, Nicolaou S, Liang T. The Evolving Function of the Radiologist: The Vancouver Workload Utilization Analysis Examine. Journal of the American School of Radiology. 2013 Oct 1;10(10):764–9.
(2) Schemmel A, Lee M, Hanley T, Pooler BD, Kennedy T, Discipline A, et al. Radiology Workflow Disruptors: A Detailed Evaluation. Journal of the American School of Radiology. 2016 Oct 1;13(10):1210–4.
(3) https://www.doximity.com/stories/physician-compensation-report/2020
(4) Dettmers, Tim, Artidoro Pagnoni, Ari Holtzman, and Luke Zettlemoyer. “Qlora: Environment friendly finetuning of quantized llms.” Advances in Neural Info Processing Methods 36 (2024).
(5) https://journal.sebastianraschka.com/p/lora-and-dora-from-scratch
[ad_2]